A blood test detecting a key protein produced by cancer cells is showing promise as an early indicator of cancer. The study, published in Cancer Discovery, September 12, suggests that the ultra-sensitive assay for LINE-1-ORF1p offers potential as an effective ‘binary’ methodology for the early detection of multiple cancers and for monitoring response to treatment.
“These kinds of ultrasensitive detection instruments are poised to improve patient outcomes in transformative ways,” says Michael Rout*, one of the authors, from the Rockefeller University, New York.
While advances have been made in detecting circulating tumour DNA, circulating tumour cells, micro RNAs and extracellular vesicles as non-invasive cancer biomarkers, achieving affordable, scalable, clinical grade screening assays has proved challenging. Although these biomarkers are expressed in higher quantities in cancer patients, one difficulty has been that they are also expressed in normal tissues, leading to a lack of specificity. The protein, Long Interspersed Element-1 (LINE-1) open reading frame 1 protein (ORF1p), offers potential as a highly specific cancer biomarker.
LINE-1 is a retrotransposon, a virus like element normally expressed in sperm and egg during embryogenesis. “Retrotransposons serve no purpose. They are in effect parasites that happen to break free when repressive silencing by methylation is wiped out early in meiosis as part of epigenetic reprogramming ensuring we are born ‘young’. Releasing LINE1 appears to be a necessary evil in this process,” Martin Taylor, the first author, tells Cancerworld. In the embryo, LINE 1 replicates through a ‘copy and paste’ mechanism, leading to a new copy in a new position in the genome. It is unknown, says Taylor, how many embryos are aborted due to damage from such insertions.
In normal somatic tissues LINE-1 expression is repressed, resulting in either very low or undetectable levels. Epigenetic dysregulation of LINE-1 and L1 ORF1p overexpression begins early in carcinogenesis, making ORF1p a promising highly specific cancer biomarker. “Most assays target a normal human protein that is overexpressed in cancer but has normal expression in tissue. LINE-1, however, is only expressed in disease, making it a uniquely specific binary marker,” explains Taylor, from Harvard Medical School.
In an earlier paper published in American Journal of Pathology in 2014, Taylor and colleagues showed LINE-1-ORF1p to be a hallmark of many cancers, including breast, liver, colon, ovary, uterus, head and neck, lung, stomach, oesophagus, pancreas, kidney, gallbladder and prostate. While brain and blood cancers also express LINE-1-ORF1p, levels are found in far lower concentrations.
Conventional ELISA analytical biochemistry assays have not been sensitive enough to detect ORF1p, so the team developed ultrasensitive digital immunoassays capable of detecting mid-attomolar (10-17 M) ORF1p concentrations from as little as 25 µL of plasma. The current study uses a single-molecule-based detection technology, known as single molecule arrays (SIMOA), which utilises nanobodies – special types of antibodies representing the smallest naturally occurring antigen-binding fragments. The technology, developed by David Walt from Brigham and Women’s Hospital, coats very small beads with nanobodies (taken from llamas) that have been customised to capture ORF1p. A second ‘detection’ antibody is added that binds to a second site on the ORF1p target protein, providing a quantifiable fluorescent signal. “The immunocomplex, consisting of bead, bound protein, and detection antibody, is around 1,000 times more sensitive than conventional ELISA, enabling us to capture proteins at really low levels, akin to finding a ‘needle in a haystack’,” eplains Taylor.
For the current study, the team investigated plasma taken from healthy people and patients with a range of cancers. First, they analysed plasma from 450 healthy people, aged 20 to 90, who had donated blood to the Mass General Brigham Biobank. ORF1p was undetectable in 97–99% of them, and of the five people with detectable ORF1p, the person with the highest level was found six months later to have advanced prostate cancer. Detectable ORF1p was seen in one of 30 patients with chronic liver disease who was subsequently diagnosed with hepatocellular carcinoma.
ORF1p was found to be more prevalent than previously appreciated in cancers of the colon and oesophagus. In colon cancer, more than 90% of 191 patients had detectable ORF1p, and in oesophageal cancer 100% of patients had detectable ORF1p.
The team demonstrated that ORF1p is an early indicator of chemotherapeutic response in gastric and oesophageal cancers at timepoints as short as 26 days (where other parameters are often ambiguous), opening the possibility to monitor minimal residual disease or relapse.
In one part of the study, the team assessed 19 patients receiving treatment for gastro oesophageal cancer. In the 13 patients who responded to therapy, levels of ORF1p fell below the detection limit of the assay. “These results suggest that the marker can be an early indicator of response in a disease where it is hard to know if the therapy is working,” says Taylor.
The assays are cost-effective (each test costs less than $3 in consumables), rapid (take less than two hours), and simple to perform. A limitation of the approach is that results do not provide key pieces of information, such as the location of the cancerous tissue in the body.
The team are currently looking to increase the sensitivity of the test so that it can detect ORF1p in blood taken from patients with brain and blood cancers, and are also looking to develop the assays for use in other biofluids, such as stool, pap smears, and urine. Ultimately, the team believe the population could have their ORF1p protein levels measured when healthy to establish a baseline. Any subsequent ‘spikes’ would then be a signal to trigger deeper investigations.
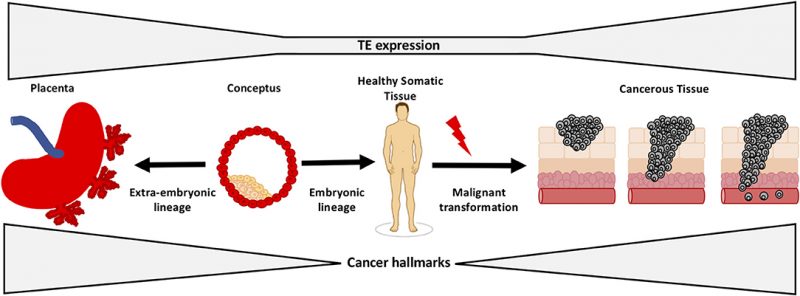
The opening illustration shows how transposons (or transposable elements “TEs”) regulate early development and tumorigenesis. It first appeared in: Lynch-Sutherland CF, Chatterjee A, Stockwell PA, Eccles MR and Macaulay EC (2020) Reawakening the Developmental Origins of Cancer Through Transposable Elements. Front Oncol 10:468. doi: 10.3389/fonc.2020.00468 It is republished under the terms of the Creative Commons Attribution License (CC BY). ©2020 Lynch-Sutherland, Chatterjee, Stockwell, Eccles and Macaulay
*The corresponding authors on the paper were David Walt (Harvard University), Kathleen Burns (Harvard Medical School) and Connie Wu (Harvard University).
This article was amended to reflect the full list of authors of this paper.