Future prospects for tackling aggressive prostate cancer emerged from two presentations at the American Association for Cancer Research Annual meeting, held virtually in mid-April. One identified how the risk of prostate cancer progressing to lethal disease might be mitigated by following healthy lifestyles. The other explored how it may be possible to identify those men with early prostate cancer who are at greatest risk of progression, through screening biopsy specimens for NKX3.1 polymorphisms (unpublished findings from a research project on Mitochondrial and nuclear functions of NKX3.1 in regulating oxidative stress in prostate cancer)1.
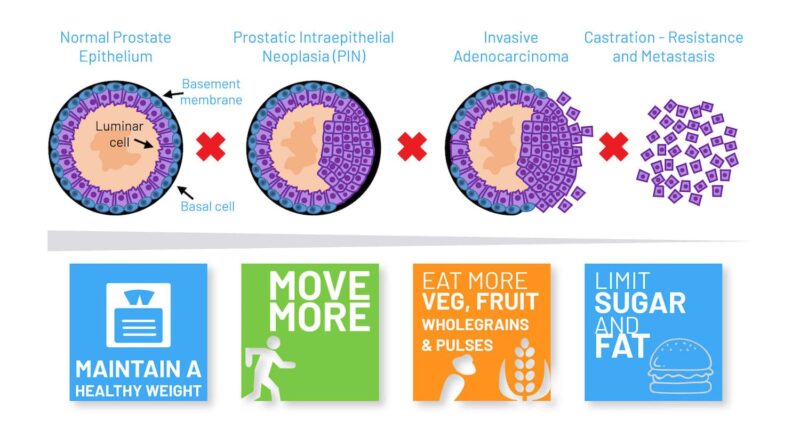
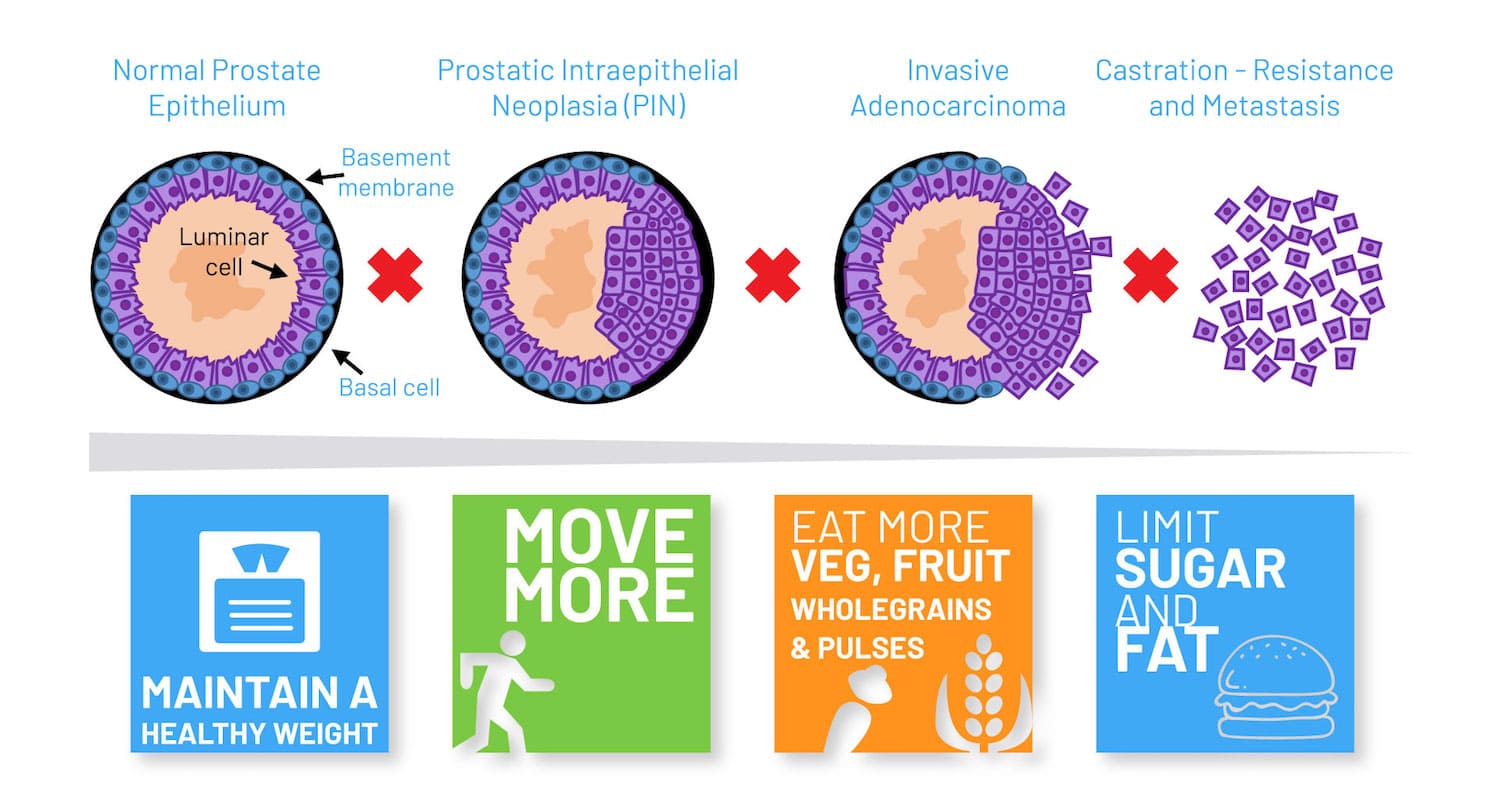
In prostate cancer, around 15% of men have high risk disease with the potential to progress to a lethal form of the disease that can prove fatal. A fundamental challenge facing clinicians is how to distinguish those with high risk tumours from those with low risk tumours, where active surveillance can be used to avoid or delay treatment.
In the first study, Anna Plym, from Brigham and Women’s Hospital and Harvard TH School of Public Health, and colleagues, explored whether men with increased genetic risk of prostate cancer might offset their risk of developing aggressive disease through adopting healthy lifestyles.
The team used a 269 polygenic risk score to quantify genetic risk of prostate cancer for 10,443 men from the Health Professionals Follow-Up study with genotype data available. The Harvard initiative, which started in 1986, aims to evaluate hypotheses around men’s health relating to nutrition.
Plym and colleagues applied lifestyle scores to their subjects, where a score of 1 to 2 indicated the least healthy lifestyle, 3 indicated a moderately healthy lifestyle, and 4 to 6 the most healthy lifestyle. Lifestyle scores were calculated using information based on weight, smoking, physical activity and diet (consumption of tomatoes, fatty fish and reduced intakes of processed meat).
The team identified 2,111 prostate cancer cases in total during a median follow-up period of 18 years, and 238 lethal prostate cancer cases during a median follow-up of 22 years.
Results showed men in the highest quartile by polygenic risk score were almost five and a half times more likely to develop prostate cancer, and three and a half times more likely to develop lethal prostate cancer than those in the lowest quartile. However, the investigators went on to show that the risk for lethal prostate cancer in men in the highest genetic risk quartile who adhered to the healthiest lifestyle was almost half that among men with an equivalent genetic risk with the least healthy lifestyle (HR 0.54; 95% CI, 0.31-0.93).
This risk reduction associated with healthier lifestyles applied specifically to risks of developing lethal prostate cancer. By contrast, no impact was shown for the risk of developing prostate cancer per se. Men in the highest risk quartile adhering to the healthiest diet did not show any difference in risk compared to men in the highest risk quartile with the least healthy diet (HR 1.01; 95% CI. 0.84-1.22).
In the high-risk group, having a healthy lifestyle at the time of study entry was associated with a lifetime cumulative risk of developing lethal prostate cancer of 3% compared with 6% among men at high risk with the least healthy lifestyle.
Commenting on the presentation, session moderator Charles Swanton, from the Francis Crick Institute and UCL Cancer Institute, London, said, “So the question is, why did a healthy lifestyle only protect those in the highest PRS [polygenic risk score] category? And so I think we need future validation in larger cohorts using similar thresholds and a biological mechanism that might explain an interaction between the healthy lifestyle and a highest genetic risk and the risk of lethal prostate cancer.”
“Men at high genetic risk may benefit from a targeted screening programme, aimed at detecting a potentially lethal prostate cancer while it is still curable”
Plym added, “Our findings add to current evidence suggesting that men with a high genetic risk may benefit from a targeted prostate cancer screening programme, aiming at detecting a potentially lethal prostate cancer while it is still curable.”
One possibility might be to screen men with early prostate cancer for NKX3.1 genetic polymorphism, suggested Cory Abate Shen in a session on Molecular Targets of Precision Prevention and Interception. It has been known for over two decades that NKX3.1 ‒ located on a key region of chromosome 8p21 ‒ is lost in 70% of early prostate cancers, explained Abate Shen, from Columbia University, New York. In addition, loss of Nkx3.1 function in mutant mice results in defective prostate development and predisposes to prostate cancer. Recently, Abate Shen and her team have discovered a novel role for the NKX3.1 gene as a mitochondrial transcription factor during oxidative stress, where it directly regulates mitochondrially encoded genes.
Abate Shen described how, when the group first compared wild-type and mutant NKX3.1 mice treated with paraquat (inducing oxidative stress), they found that mutant mice lacking the gene developed higher grade prostatic intraepithelial neoplasia (PIN) phenotypes. One intriguing observation was that NKX3.1 responded to oxidative stress by becoming localised to both cell nuclei and mitochondria. When the team explored the functions of proteins encoded by common NKX3.1 polymorphisms that are associated with risk of aggressive cancer, they found that, following oxidative stress, the mutated proteins were either unable to be localised to the mitochondria or could not bind to mitochondrial DNA. Further investigations showed that the mitochondrial chaperone protein HSAP9 was required for NKX3.1 transport into the mitochondria to provide protection against reactive oxygen species, and that NKX3.1 bound to the D Loop of mitochondrial DNA.
To investigate NKX3.1’s role in primary prostate cancer, the team developed organotypic cultures using prostate tissue collected from surgery. Building on earlier observations that patients with prostate cancer could have both high and low NKX3.1 expression, the team explored how NKX3.1 levels corresponded with real world outcomes of patients providing biopsy samples. Results showed patients with low NKX3.1 and low mitochondria NKX3.1 gene expression had the worst outcomes; while those high NKX3.1 and high NKX3.1 mitochondria gene expression had the best outcomes.
“It is plausible that, simply by staining for NKX3.1 in biopsy samples, we could identify those patients who are most at risk of progressing”
Taken together, says Abate Shen, the data demonstrate a new mechanism for prostate cancer suppression through NKX3.1, with NKX3.1 polymorphisms that encode proteins that cannot respond to oxidative stress representing one means by which men are prone to develop more aggressive forms of prostate cancer. “Quite strikingly, the data suggests both expression levels and subcellular location of NKX3.1 are associated with disease outcome as well as expression of the mitochondrial genes being regulated,” said Abate Shen. “It is plausible that, simply by staining for NKX3.1 in biopsy samples, we could identify those patients who are most at risk of progressing.”
Taken together these two studies point the way to a plausible strategy for reducing the incidence of lethal prostate cancer through identifying those cases of prostate cancer that carry a genetic high risk of developing lethality, and helping men identified with such disease to reduce that risk through adopting healthy lifestyles.
1. Some of the precursor work done by Abate Shen and her colleagues on the role of NKX3.1 in prostate cancer can be found in C Le Magnen et al (Dis Model Mech 2018) and A Dutta et al (Eur Urol 2017).